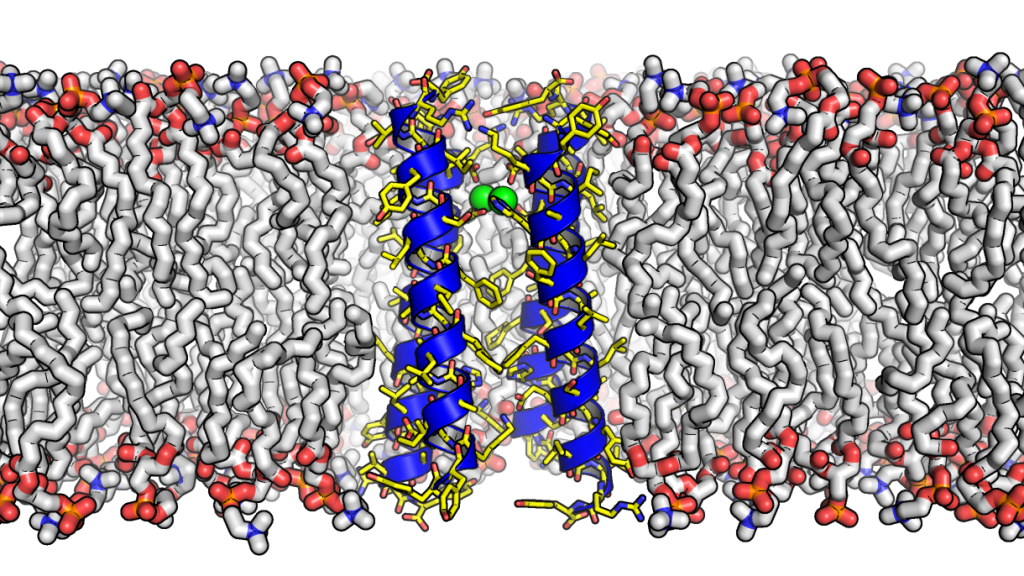
Human cells are protected by a largely impenetrable molecular membrane, but Prof. Gevorg Grigoryan, with a team of collaborators, have built the first artificial transporter protein that carries individual atoms across membranes, opening the possibility of engineering a new class of smart molecules with applications in fields as wide ranging as nanotechnology and medicine. This work, which appeared in the journal Science, is a milestone in designing and understanding membrane proteins. The study was a collaboration between researchers across different universities: Dartmouth's Gevorg Grigoryan, MIT's Mei Hong, fellow University of California, San Francisco investigators William F. DeGrado and Michael Grabe, and others.
Each human cell is surrounded by a lipid membrane, a molecular barrier that serves to contain the cellular machinery and protect it from the surrounding elements. This cellular “skin” is impenetrable to most biological molecules but also presents a conundrum: if chemicals can’t get in our out, how is a cell to receive nutrients (food) and remove unwanted products of metabolism (trash)? Nature has come up with an elegant solution to this logistical problem — transporter proteins (or transporters). These molecular machines are embedded in the cellular membrane and serve as gatekeepers, allowing specific chemicals to shuttle in and out when needed. Though biologists have known about transporters for many decades, their precise mechanism of action has been elusive.
The famous physicist Richard P. Feynman once said, “What I cannot create, I do not understand.” Following this logic, the researchers set out to “build” an artificial transporter protein from scratch, to learn how transporters work and to open the possibility of engineering a new class of smart molecules. They developed new computational techniques to model the necessary molecular physics, enabling them to design a transporter protein through computer simulation. Specifically, computer simulations suggested which amino-acid building blocks should comprise the future transporter, so that it would carry ionic atoms of metal zinc in one direction across membranes, while pumping protons in the other. Using this computational blueprint, they created the molecule in the lab, referring to it as “Rocker” due to its predicted molecular dynamic properties: the protein was expected to “rock” between two alternating states, allowing it to drive atoms through.
“To our great excitement, experiments showed that Rocker did indeed transport zinc and protons and it did, in fact, rock between two states just as designed,” says co-lead author Gevorg Grigoryan, an assistant professor of computer science and adjunct assistant professor of biological sciences at Dartmouth. “Further, Rocker showed great selectivity, not transporting ions of calcium, another design feature.”
Proteins are nature’s workhorse molecules, performing a great variety of tasks in the cell from catalysis and sensing to generation of mechanical work. Learning to design (from first principles) novel protein molecules to perform specific tasks would mean that the immense richness of function that proteins have to offer can be brought to bear in a variety of applications — from better therapeutics to smart materials and clean energy solutions.
“Our findings are an important step forward in this pursuit, demonstrating that through the use of computer simulations to orchestrate precise properties of atomic structure and molecular dynamics, proteins can now be designed to carry out complex functions that rival those of natural molecular machines,” Grigoryan says. Dr. Andrei N. Lupas of Max Planck Institute for Developmental Biology, a leading expert in protein design, has a similar assessment of the work, writing in a perspective article that the study "convincingly breaks several barriers in protein engineering: taking protein design from the solvent into the membrane, aiming for dynamic properties rather than for stability, and achieving an advanced biomimetic function from first principles, without recourse to screening or directed evolution."
Dartmouth Assistant Professor Gevorg Grigoryan is available to comment at gevorg.grigoryan@dartmouth.edu.